-Search query
-Search result
Showing 1 - 50 of 84 items for (author: taylor & ms)

EMDB-40967: 
Cryo-EM structure of a full-length, native Drp1 dimer
Method: single particle / : Rochon K, Mears JA

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
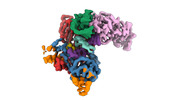
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
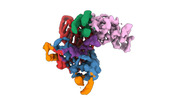
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-40789: 
BAP1/ASXL1 bound to the H2AK119Ub Nucleosome
Method: single particle / : Thomas JF, Valencia-Sanchez MI, Armache KJ

EMDB-40790: 
Map focused on acidic patch BAP1/ASXL1 bound to the H2AK119Ub Nucleosome
Method: single particle / : Thomas JF, Valencia-Sanchez MI

EMDB-40791: 
Overall map of BAP1/ASXL1 bound to the H2AK119Ub Nucleosome
Method: single particle / : Thomas JF, Valencia-Sanchez MI

PDB-8svf: 
BAP1/ASXL1 bound to the H2AK119Ub Nucleosome
Method: single particle / : Thomas JF, Valencia-Sanchez MI, Armache KJ

EMDB-26735: 
Hantavirus ANDV Gn(H) protein in complex with 2 Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE
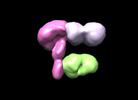
EMDB-26736: 
Hantavirus MAPV Gn(H)/Gc protein in complex with 2 Fabs SNV-24 and SNV-53
Method: single particle / : Binshtein E, Crowe JE

EMDB-27318: 
CryoEM structure of Hantavirus ANDV Gn(H) protein complex with 2Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE

EMDB-27178: 
Structure of Cas12a2 binary complex
Method: single particle / : Bravo JPK, Taylor DW
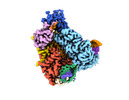
EMDB-27179: 
Cas12a2 quaternary complex
Method: single particle / : Bravo JPK, Taylor DW
EMDB-27180: 
Structure of Cas12a2 ternary complex
Method: single particle / : Bravo JPK, Taylor DW

EMDB-27061: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-57
Method: single particle / : Huang W, Taylor D

EMDB-27067: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 1-22
Method: single particle / : Huang W, Taylor D

EMDB-27068: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-65
Method: single particle / : Huang W, Taylor D

EMDB-27069: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-34
Method: single particle / : Huang W, Taylor D

EMDB-27071: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 1-23
Method: single particle / : Huang W, Taylor D

EMDB-27072: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-67
Method: single particle / : Huang W, Taylor D

EMDB-27073: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 1-8
Method: single particle / : Huang W, Taylor D

EMDB-27074: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-34
Method: single particle / : Huang W, Taylor D

EMDB-27075: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-45
Method: single particle / : Huang W, Taylor D

EMDB-27080: 
RBD of SARS-CoV-2 Spike protein in complex with pan-sarbecovirus nanobodies 2-10, 2-67, 2-62 and 1-25
Method: single particle / : Huang W, Taylor D

PDB-8cxn: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-57
Method: single particle / : Huang W, Taylor D

PDB-8cxq: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 1-22
Method: single particle / : Huang W, Taylor D

PDB-8cy6: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-65
Method: single particle / : Huang W, Taylor D

PDB-8cy7: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-34
Method: single particle / : Huang W, Taylor D

PDB-8cy9: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 1-23
Method: single particle / : Huang W, Taylor D

PDB-8cya: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-67
Method: single particle / : Huang W, Taylor D

PDB-8cyb: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 1-8
Method: single particle / : Huang W, Taylor D

PDB-8cyc: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-34
Method: single particle / : Huang W, Taylor D

PDB-8cyd: 
SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-45
Method: single particle / : Huang W, Taylor D

PDB-8cyj: 
RBD of SARS-CoV-2 Spike protein in complex with pan-sarbecovirus nanobodies 2-10, 2-67, 2-62 and 1-25
Method: single particle / : Huang W, Taylor D

EMDB-24833: 
Cryo-EM structure of Cas9 in complex with 12-14MM DNA substrate, 5 minute time-point
Method: single particle / : Bravo JPK, Taylor DW, Liu MS, Johnson KA

EMDB-24834: 
Cas9 in complex with 12-14MM DNA at 60 minute time-point, linear conformation
Method: single particle / : Bravo JPK, Taylor DW, Liu MS, Johnson KA

EMDB-24835: 
Cas9 bound to 12-14MM DNA, 60 min time-point, kinked conformation
Method: single particle / : Bravo JPK, Taylor DW, Liu MS, Johnson KA

EMDB-24836: 
Cas9 in complex with 15-17MM DNA, 60 min time-point
Method: single particle / : Bravo JPK, Taylor DW, Liu MS, Johnson KA

EMDB-24837: 
Cas9 in complex with 18-20MM DNA, 1 min time-point, linear conformation
Method: single particle / : Bravo JPK, Taylor DW, Liu MS, Johnson KA

EMDB-24838: 
Cas9:gRNA in complex with 18-20MM DNA, 1 minute time-point, kinked active conformation
Method: single particle / : Bravo JPK, Taylor DW, Liu MS, Johnson KA

EMDB-23379: 
Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S7D
Method: single particle / : Andreas MP, Adamson L, Tasneem N, Close W, Giessen T, Lau YH

EMDB-23380: 
Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S1K
Method: single particle / : Andreas MP, Adamson L, Tasneem N, Close W, Giessen T, Lau YH

EMDB-23381: 
Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S1R
Method: single particle / : Andreas MP, Adamson L, Tasneem N, Close W, Giessen T, Lau YH

EMDB-23382: 
Thermotoga maritima Encapsulin Nanocompartment Pore Mutant SE3
Method: single particle / : Andreas MP, Adamson L, Tasneem N, Close W, Giessen T, Lau YH

EMDB-23383: 
Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S6E
Method: single particle / : Andreas MP, Adamson L, Tasneem N, Close W, Giessen T, Lau YH

EMDB-23384: 
Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S5D
Method: single particle / : Andreas MP, Adamson L, Tasneem N, Close W, Giessen T, Lau YH

EMDB-23385: 
Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S7G
Method: single particle / : Andreas MP, Adamson L, Tasneem N, Close W, Giessen T, Lau YH
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
